Object ccGenomicLink will call the function circlize::circos.genomicLink while drawing.
Arguments
- region1
A data frame in bed format.
- region2
A data frame in bed format.
- rou
Pass to
circos.link.- rou1
Pass to
circos.link.- rou2
Pass to
circos.link.- col
Pass to
circos.link, length can be either one or nrow ofregion1.- lwd
Pass to
circos.link, length can be either one or nrow ofregion1.- lty
Pass to
circos.link, length can be either one or nrow ofregion1.- border
Pass to
circos.link, length can be either one or nrow ofregion1.- ...
Pass to
circos.link.
Value
Object ccGenomicLink
Examples
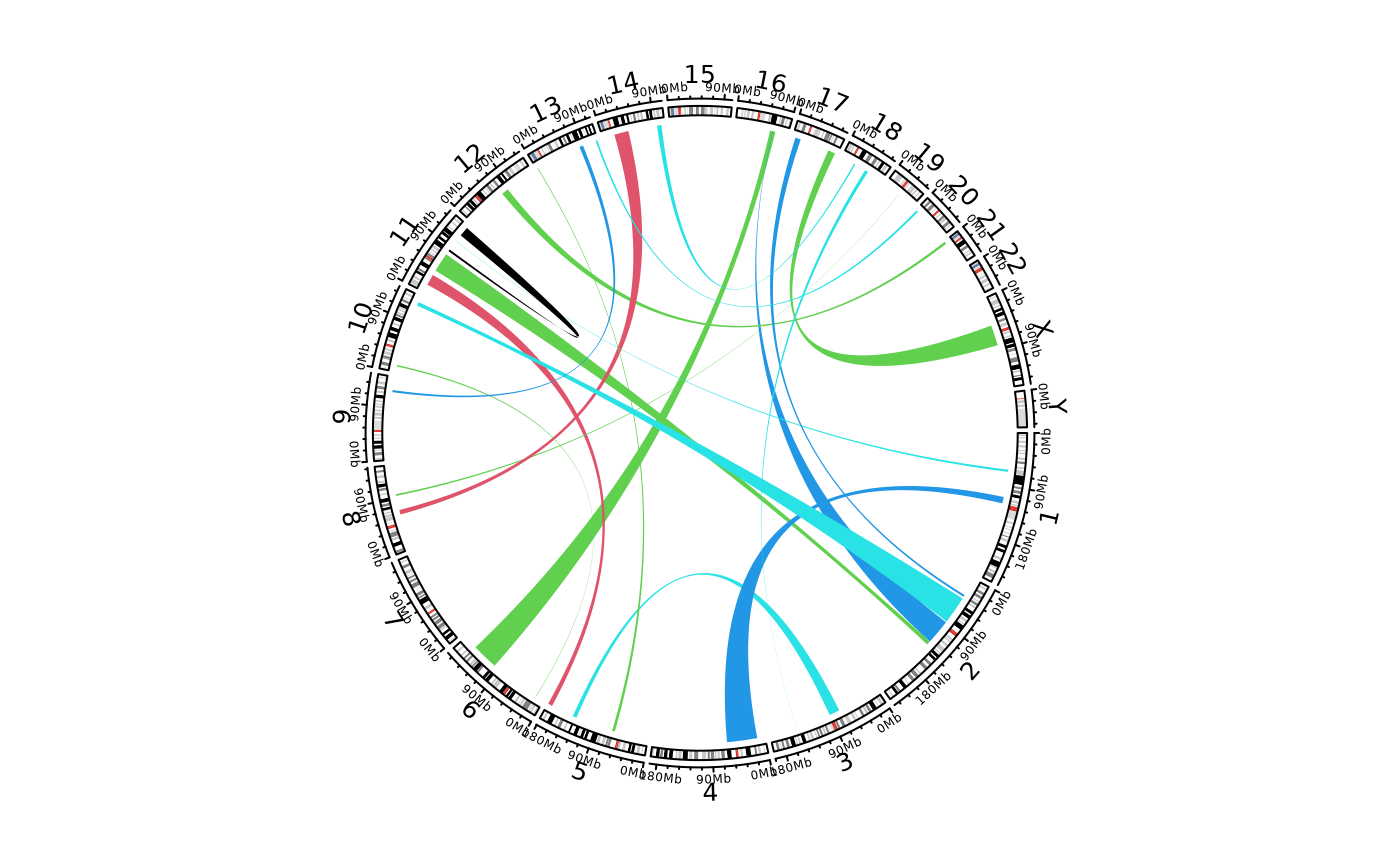
# \donttest{
library(circlizePlus)
set.seed(123)
bed1 = generateRandomBed(nr = 100)
bed1 = bed1[sample(nrow(bed1), 20), ]
bed2 = generateRandomBed(nr = 100)
bed2 = bed2[sample(nrow(bed2), 20), ]
par1 = ccPar("track.height" = 0.1, cell.padding = c(0, 0, 0, 0))
cc = ccPlot(initMode="initializeWithIdeogram")
link1 = ccGenomicLink(bed1, bed2, col = sample(1:5, 20, replace = TRUE), border = NA)
cc + par1 + link1
 # }
# }