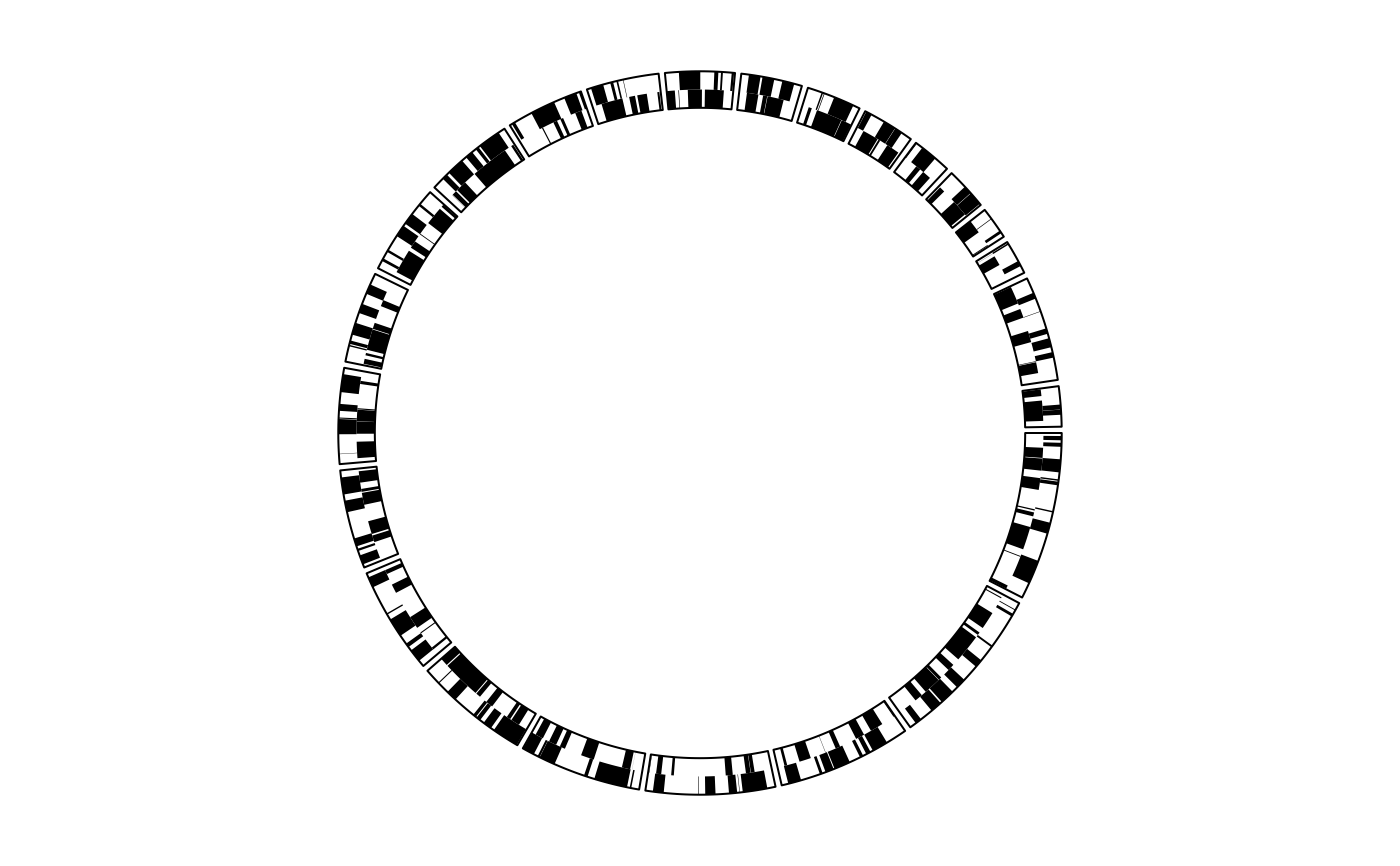
Object ccGenomicCellGeom will call the function circlize::circos.genomicRect while drawing.
Usage
ccGenomicRect(
region = NULL,
value = NULL,
ytop = NULL,
ybottom = NULL,
ytop.column = NULL,
ybottom.column = NULL,
posTransform = NULL,
col = NA,
border = "black",
lty = par("lty"),
...
)Arguments
- region
A data frame contains 2 column which correspond to start positions and end positions.
- value
A data frame contains values and other information.
- ytop
A vector or a single value indicating top position of rectangles.
- ybottom
A vector or a single value indicating bottom position of rectangles.
- ytop.column
If
ytopis invalue, the index of the column.- ybottom.column
If
ybottomis invalue, the index of the column.- posTransform
Self-defined function to transform genomic positions, see
posTransform.defaultfor explaination.- col
The length of
colcan be either one or number of rows ofregion. Pass tocircos.rect.- border
Settings are similar as
col. Pass tocircos.rect.- lty
Settings are similar as
col. Pass tocircos.rect.- ...
Mysterious parameters.
Value
Object ccGenomicCellGeom
Examples
library(circlizePlus)
par1 <- ccPar("track.height" = 0.1, cell.padding = c(0, 0, 0, 0))
cc <- ccPlot(initMode = "initializeWithIdeogram", plotType = NULL)
bed1 <- generateRandomBed(nr = 100)
bed2 <- generateRandomBed(nr = 100)
bed_list <- list(bed1, bed2)
f <- colorRamp2(breaks = c(-1, 0, 1), colors = c("green", "black", "red"))
track1 <- ccGenomicTrack(data = bed_list, stack = TRUE)
all_chr <- c("chr1", "chr2", "chr3", "chr4", "chr5", "chr6", "chr7", "chr8",
"chr9", "chr10", "chr11", "chr12", "chr13", "chr14", "chr15", "chr16",
"chr17", "chr18", "chr19", "chr20", "chr21", "chr22", "chrX", "chrY")
rect1 <- ccGenomicRect(col = 1, border = NA)
cells1 <- ccCells(sector.indexes = all_chr) + rect1
cc + par1 + (track1 + cells1)